
Comparing results from actual recorded ancestry, to that predicted by Ancestry.com, 23andme, My Heritage, Living DNA, FT-DNA and more.
Recorded Ancestry
Results


Comparing results from actual recorded ancestry, to that predicted by Ancestry.com, 23andme, My Heritage, Living DNA, FT-DNA and more.





But I'm afraid I don't buy it.


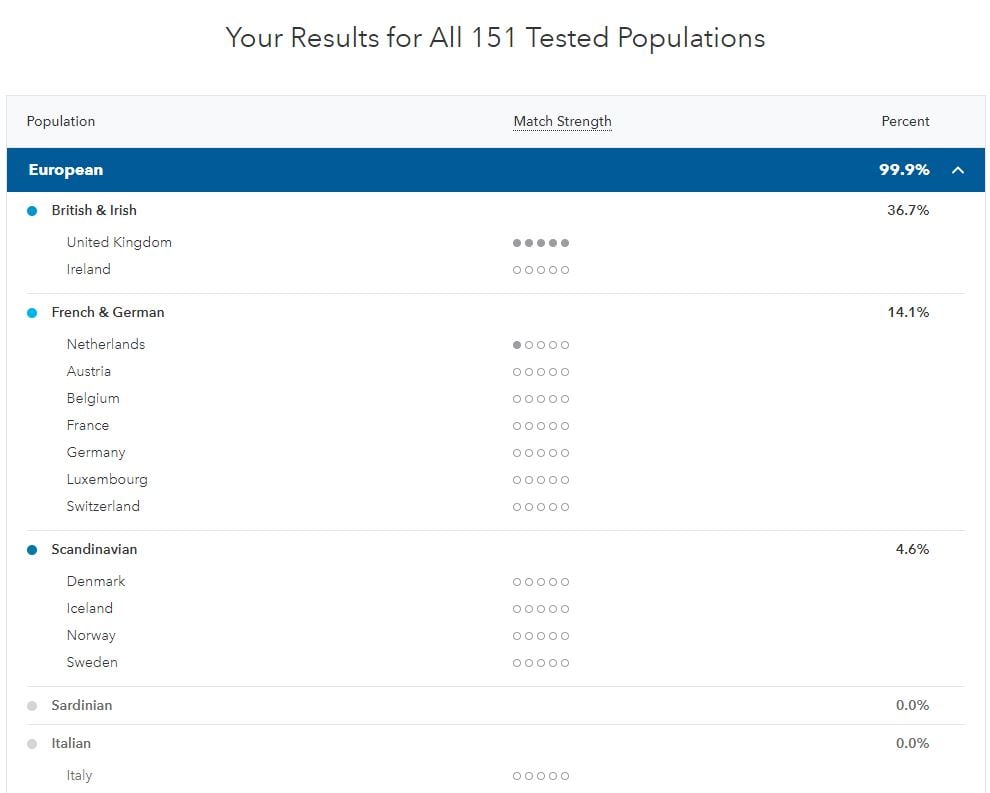
Therefore in summary - I've come to the conclusion that NO current autosomal DNA test for ancestry is capable of accurately predicting your ancestry below a very large region, such as NW Europe - unless your ancestors belong to a particularly well defined population that avoided medieval admixture. They are all inaccurate. More important to me now is that they have fat databases of testers, with a system of searching them alongside family trees, ancestor locations and surnames. However, side by side - for my results, and weighed simply against recorded family history, I have to pronounce AncestryDNA/.com to be more accurate than 23andme.

Above image. My Global 10 Genetic Map coordinates: PC1,PC2,PC3,PC4,PC5,PC6,PC7,PC8,PC9,PC10 ,0.019,0.0272,0.0002,-0.0275,-0.0055,0.0242,0.0241,-0.0033,-0.0029,0.0015. The cross marks my position on a genetic map by David Wesolowski, of the Eurogenes Blog.
The above map shows genetic distances between different human populations around the planet. Look how tightly the Europeans cluster. Razib Kahn recently blogged on just this subject. The fact of the matter is that the greatest diversity exists between populations outside of Europe, particularly within Africa, and between African and non-African populations. However, we obsess over tiny differences within European populations, when in truth, most Western Eurasians are very closely related. We share ancient ancestry from slightly varied mixes of only three base ancestral groups, with the last layer arriving only 4,300 years ago. This obsession in the Market drives DNA to the consumer businesses to largely ignore non-European diversity, and to focus too closely on differences that blur into each other.

The above image is from CARTA lecture. 2016. Johannes Krause of the Max Planck Institute. It shows the currently three known founder populations of Europeans and their average percentages.
However, at the same time the new Living DNA service seeks to zoom in closer on British populations, attempting to detect ancestry percentages from such tiny zones as "East Anglia". They appear to be having a level of success with it as well, although that blurriness, that overlap and closeness of populations in Europe gives problems. Germans are given false percentages of British, Some Scottish appear as Northern Irish, and the Irish dilute into false British areas. However, I've seen enough results now to suggest that it is far from genetic astrology. They get it correct to a certain level, particularly for us with English ancestry. Ancestry DNA customers expect perfection. I don't think that we will ever get that from such closely related populations at this resolution, but it does provide a new genealogical tool that can point us into some revealing directions.

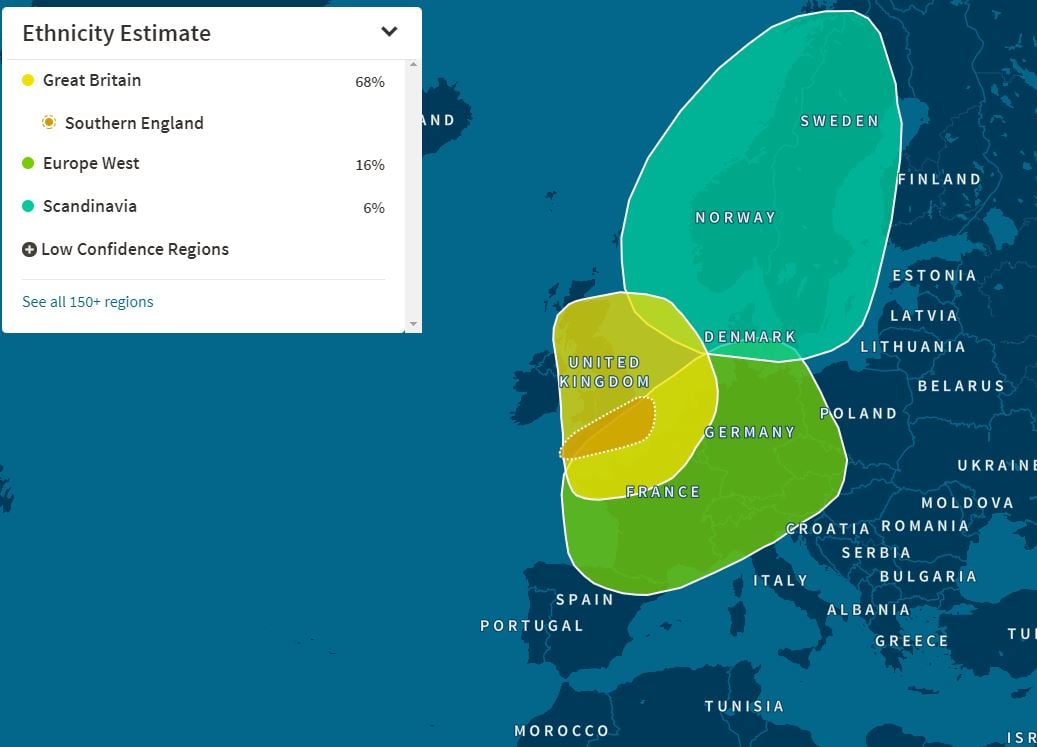
Above image. My Living DNA Map. Based on my recorded genealogy, I estimate 77% to 85% East Anglian ancestry over the past 250 years or so. Living DNA at Standard Mode gave me 39%. I'm impressed by that. That a DNA test can recognise even at a 50% success, my recent ancestry in such a tiny zone of the planet. I have doubts though that this sort of test will ever be free of errors, and mistakes. The safest DNA test for ancestry is still one that is based on more distinct populations, and outside of Africa, that can be as wide as "European". 23andMe for example in their "Standard Mode" (75% confidence), assign me 97.3% European, and 0.3% Unassigned. That is a pretty safe result.
Autosomal DNA tests for ancestry, particularly for West Eurasian (European and Western Asia) descendants, are not reliable at high resolution. If you want to get really local, then sure - do it. However, only use the results as an indication, not as a truth. Populations in Western Eurasia are closely related, and share recent common descent. There has been a high degree of mobility and admixture ever since. Some modern populations tested do not have a high level of deep rooted local ancestry in that region. They overlap with each other. Keep researching and meander through different perspectives of what your older pre-recorded ancestry could have been.

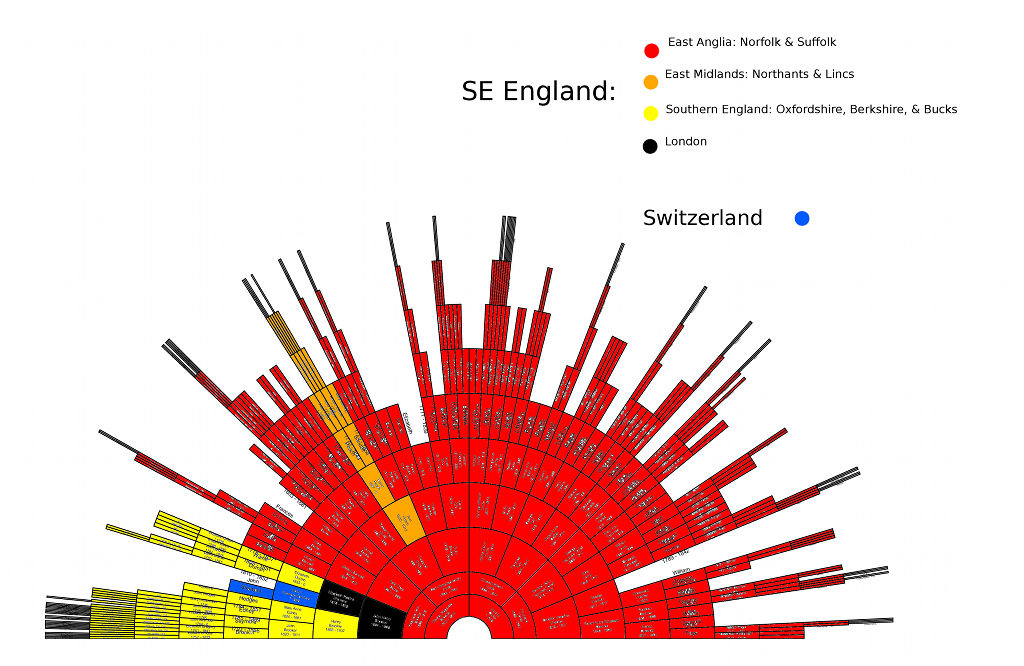
Above image by Anthrogenica board member Tolan. Based on 23andMe AC results. My results skew away from British, and towards North French. He generated this map, plotting myself (marked as Norfolk in red), and my Normand Ancestral DNA twin Helge in yellow. My results fall in the overlap with French. Helge is Normand but in AC appears more British than myself. I am East Anglian yet in this test appear more French than he does.

I have taken several DNA tests for ancestry, including those provided by the FT-DNA, 23andMe, and Living DNA companies. Unusual for a tester, I am actually of a single population, very local, well documented ancestry here in East Anglia, South-East England. I'm not someone in the Americas or Australia, that might have very little clue what parts of the world that their ancestors lived in, previous to immigration. I know my roots, I'm lucky. I live them. You might ask, why did I feel the need to test DNA for ancestry? The answer is, curiosity, to test the documented evidence, fill the gaps, look for surprises, and in particular, to understand the longer term, to reach further back into my ancestry.
I have though, become a bit of a skeptic, even a critic, of autosomal DNA (auDNA) tests for ancestry. They are the tests presented by the businesses in results called something like Ancestry, Family Ancestry, Origins, Family, Composition, etc. Instead of testing the haplogroups on either the direct paternal (Y-DNA), or direct maternal (mtDNA), these tests scan the autosomal and X chromosomes. That's good, because that is where all of the real business is, what makes you an individual. However, it is subject to a phenomena that we call genetic recombination (the X chromosome is a little more complicated). This means that every generation circa 50% of both parents DNA is randomly inherited from each parent. I said randomly. Each generation, that randomness chops up the inherited segments smaller, and moves them around. After about seven or eight generations, the chances of inheriting any DNA from any particular ancestral line quickly diminishes. It becomes washed out by genetic recombination.

Therefore, not only are the autosomes subject to a randomness, and genetic recombination - they are only useful for assessing family admixture only over the past three hundred years or so. There is arguably, DNA that has been shared between populations much further back, that we call background population admixture. It survived, because it entered many lines, for many families, following for example, a major ancient migration event. If this phenomena is accepted - it can only cause more problems and confusion, because it can fool results into suggesting more recent family admixture - e.g. that a great grandparent in an American family must have been Scandinavian, when in fact many Scandinavians may have settled another part of Europe, and admixed with that ancestral population, more than one thousand years ago.

DNA businesses compare segments of auDNA, against those in a number of modern day reference populations or data sets from around the world. They look for what segments are similar to these World populations, and then try to project, what percentages of your DNA is shared or similar to these other populations. Therefore:

It is far truer to say that your auDNA test results reflect shared DNA with modern population data sets, rather than to claim descent from them. For example, 10% Finnish simply means that you appear to share similar DNA with a number of people that were hopefully sampled in Finland (and hopefully not just claim Finnish ancestry) - not that 10% of your ancestors came from Finland. That is, for the above reasons, presumptuous. It might indeed suggest some Finnish ancestry, but this is where many people go wrong, it does not prove ancestry from anywhere.
This is my main quibble. So many testers take their autosomal (for Family/Ancestry) DNA test results to be infallible truths. They are NOT. White papers do not make a test and analysis system perfect and proven as accurate. Regarding something as Science does not make it unquestionable - quite the opposite. The fact of the matter is, if you test with different companies, different siblings, add phasing, you receive different ancestry results. Therefore which result is true and unquestionable?

So what use is DNA testing for ancestry? Actually, I would say, lots of use. If you take the results with a pinch of salt, test with different companies, then it can help point you in a direction. Never however take autosomal results as infallible. Critical is to test with companies with well thought out, high quality reference data sets. Also to test with companies that intend to progress and improve their analysis and your results.
For DNA relative matching, then sure, the companies with the best matching system, the largest match (contactable customer) databases, and with custom in the regions of the world that you hope to match with. There is also, GEDmatch. Personally, I find it thrilling when I match through DNA, but in truth, I had more genealogical success back in the days when genealogists posted their surname interests in printed magazines and directories.
The results of each ancestry test should be taken as a clue. Look at the results of testers with more proven documented and known genealogies. Learn to recognise what might be population background, as opposed to recent admixture in a family. Investigate haplogroup DNA - it has a relative truth, although over a much longer time, and wider area. Just be aware that your haplogroup/s represent only one or two lines of descent - your ancestry over the past few thousand years may not be well represented by a haplogroup. Investigate everything. Enjoy the journey. Explore World History.

I just see so many misunderstandings on genetic genealogy and DNA test forums concerning mtDNA haplogroups, that I feel compelled to try to explain.
DNA testing businesses tend to dumb down a lot of information for their "audience". I feel that this actually increases misunderstandings, and mtDNA haplogroups are a good example. Rather than use the lengthy description mitochondrial DNA, or even it's shortened mtDNA, businesses describe it more frequently as Mother Line, or Maternal. It misleads so many of their customers. So let us put this straight:
So what does this mean in practice?
That is all that I wanted to say. it is a fascinating marker, but it is not representative of even 50% of your ancestry, it is not an identity, it is pretty irrelevant to surname (studies), it is inherited only down one narrow line - but all of the way back.

My earliest mtDNA ancestor with a surviving photograph. My mother's mother's, mother's, mother (2xgreat grandmother), born Sarah Daynes in Norfolk, during 1845. Her mtDNA would be H6a1a8. Her mother was born Sarah Quantrill in Norfolk during 1827. Her mother in turn was born Mary Page in Norfolk during 1791. Her mother in turn was born Elizabeth Hardiment in Norfolk during 1751. Her mother in turn (my 6xgreat grandmother) was Susannah Briting, who married John Hardyman in Norfolk during 1747. If my documentary research along this line is correct, then Susannah inherited mtDNA haplotype H6a1a8 from her mother.

My Global 10 Genetic Map coordinates: PC1,PC2,PC3,PC4,PC5,PC6,PC7,PC8,PC9,PC10 ,0.019,0.0272,0.0002,-0.0275,-0.0055,0.0242,0.0241,-0.0033,-0.0029,0.0015
This is my position on the latest genetic map by David Wesolowski, of the Eurogenes Blog. One point of interest that has been picked up on the Anthrogenica Forums, is my consistent closeness in ancestral results, to a Normand member! Our Basal-rich K7 results were almost identical. On 23andMe Ancestry Composition (spec mode), I just get a bit more French & German, while he gets just a bit more British & Irish. We are close!
Another forum member argued though that it's my results that are skewed away from British, and towards North French. He generated this map, plotting myself (marked as Norfolk in red), and my Norman Ancestral DNA twin Helge in yellow:

I had to point out though, that I've rarely seen other SE English with a record of local ancestry, test - and that the red circles representing British & Irish include many people with some Irish, Scottish, Welsh, Western, or Northern ancestry. The map suggests a pull to Northern France, Belgium, and the Netherlands.
As I commented towards the end of my last post, I initially expected a pull to Denmark, Northern Germany, and perhaps to the Netherlands. This is because so many of my 17th-20th century ancestors lived on what was the frontier of Anglo-Saxon and Danish immigration during the 4th to 11th centuries.
But instead, autosomal DNA tests for ancestry all seem to be suggesting more shared ancestry from a more southerly direction - Northern France and Belgium particularly. Although there has so far been a dearth of local testers from local families, the POBI survey seems to find this common among the English. We appear to be a halfway house between Old British, and the French, more than the ancestors of Anglo-Saxons and Danes. This contradicts the historical and archaeological records. POBI suggested that this was due to waves of unrecorded immigration from the South during late prehistory. Others have pointed the finger at Norman and French admixture in medieval Southern Britain. It could be both!
Can I apply for French citizenship?